Biomedical Business & Technology Contributing Editor
From consolidations to point-of-care testing to nanotechnologies to analysis of cells, the diagnostics industry is downsizing — but by employing an increasing array of innovative devices and technologies
ST. LOUIS — The Oak Ridge Conference, organized here by the American Association for Clinical Chemistry (AACC, Washington) and now in its 39th year, provides a forum for presentation of the latest new technologies for clinical diagnostics and for assessment of their impact on the diagnostics market.
This year’s conference, held here in early May, covered a range of technologies, including new labeling and detection systems, point-of-care (POC) diagnostics and diagnostics for “low-resource” settings, emerging technologies for use in molecular diagnostics, including advanced sequencing technologies, and advances in cell diagnostics, including applications in which cells are being used as sensors for rapid diagnostic testing.
Structural changes are occurring in the clinical diagnostics market in addition to technology-driven changes as a result of recent acquisitions, including the acquisition of the laboratory diagnostics business of Bayer AG (Leverkusen, Germany) and Diagnostic Products Corporation (Los Angeles) by Siemens Medical Solutions (Munich, Germany); the pending acquisition of the core laboratory and POC testing business of Abbott Laboratories (Abbott Park, Illinois) by GE Healthcare (Chalfont St. Giles, UK); and the pending acquisition of Biosite (San Diego) by Inverness Medical Innovations (IMI; Waltham, Massachusetts).
These acquisitions are not likely to have significant impact on the degree of supplier consolidation in the market: the top 10 suppliers will account for 76.5% of the worldwide market, compared to 74.8% prior to the acquisitions (excluding the effect of the Biosite acquisition), but will result in a significant change in the line-up of leading companies in the market.
In particular, the No. 3 and No. 4 players in the market (GE Healthcare and Siemens) have no prior presence in clinical lab and alternate site diagnostics except for their positions in clinical information systems. Their positions in imaging and patient monitoring, where both are leaders, could have a significant impact on the market in the future, however, as both companies begin to implement a strategy integrating in vitro and in vivo diagnostics, and that also is expected to rely on a broader role for informatics.
However, the new market structure could change the basis of competition in the laboratory products market, altering the mix of technologies needed to compete effectively.
Diagnostics going ‘nano’
Nanodiagnostics, one of the newest technologies in this sector, was discussed the Oak Ridge Conference by K.K. Jain, of Jain Pharmabiotech (Basel, Switzerland), emphasizing its potential broad impact on testing, both in the central lab and at point-of-care. Jain reviewed recent developments in this sub-sector of nanomedicine, encompassing applications in in vitro diagnostics, in vivo diagnostics and imaging, and therapeutics.
Advantages of nanotechnology in diagnostics include the ability to analyze the function of cellular structures at the nanoscale level, allowing, for example, assessment of receptors and pores in cell membranes; tagging of cells with nanoparticle labels; analysis of chromosomes on a molecular scale including nanodissection and nanoextraction of chromosomal DNA; detection of unique structures on cell surfaces that have diagnostic significance; and integration of diagnostics and therapeutics at the cellular level. Nanotechnology may prove to be an enabling technology for integrating in-vitro and in-vivo diagnostics.
Jain described perfluorocarbon/lipid nanoparticles under development by Kereos (St. Louis) that can be used to label stem cells to allow tracking of cell position using MR imaging after they are injected in the body. Nanoparticle labels are also being evaluated for in vivo imaging of structures not detectable with existing imaging techniques, such as small tumor metastases, small cerebral ischemic lesions that may serve as early indicators of an impending stroke, or intracranial tumors that are not identified with existing contrast agents due to the impediment of the blood-brain barrier. Nanoparticle tags can cross the blood-brain barrier, giving them a unique capability for diagnosis of neurological disorders.
Another example of a unique application for nanotechnology is a Lipoparticle Biosensor under development by Integral Molecular (Philadelphia, Pennsylvania). Integral is developing a system employing nanoscale lipid particles used as labels in biochemical assays in continuous flow microfluidic analyzers. Such devices, due to their small reagent volume requirements, potential for rapid assay time, high sensitivity and low cost, could revolutionize central lab testing as well as POC diagnostics.
Molecular also going ‘nano’
In molecular diagnostics, nanotechnology is enabling single-molecule analysis of nucleic acids, using systems such as the Nanoanalyzer platform from BioNanomatrix (Philadelphia), which being developed for direct analysis of genomic DNA. Likewise, Lumera (Bothell, Washington) is developing protein microarrays based on its NanoCapture technology, with applications in the emerging area of proteomics. Nanotechnology allows fabrication of devices with controlled features on the molecular level, enabling design of devices with characteristics that cannot be created with conventional sensor fabrication technologies.
For example, the spacing of capture probes on a sensor substrate can be precisely controlled using nanotechnology fabrication methods, which provides improved specificity and efficiency of bonding. In addition to enhanced specificity, detection of single bacteria within a period of as little as 20 minutes may prove feasible with nanotechnology-based devices, potentially enabling real-time POC diagnosis of infectious diseases prior to starting therapy.
Oxonica (Mountain View, California) is developing the Nanoplex Rapid and Nanoplex Direct platforms for clinical diagnostics. As discussed at the conference by Michael Natan of Oxonica, the Nanoplex platform employs glass-coated nanoparticles as labels, Surface Enhanced Raman Scattering for detection, a lateral flow assay configuration, and both clinical lab and POC analyzer configurations.
The nanoparticle labels, known as SERS Nanotags, consist of layered nanostructures that provide electromagnetic amplification of the normally weak Raman optical signals to achieve a 106- to 109-fold enhancement in signal intensity, enabling very high detection sensitivity. The nanotags also exhibit very high chemical and optical stability and are insensitive to environmental factors.
The company has developed prototype assays for influenza A and B, as well as Respiratory Synctial Virus with quantitative capability and a 100-fold improvement in detection sensitivity compared to existing lateral flow immunoassay test strips. The company’s new Nanoflex Direct platform, disclosed publicly for the first time at the Oak Ridge Conference, uses magnetic particle separation to eliminate the wash steps normally required for lateral flow assays.
Oxonica has developed a cardiac troponin assay with 40 pg/mL sensitivity that requires no wash steps, no external separation steps, and that provides a single-digit coefficient of variation. A CK-MB assay has also been developed, as well as a multiplex troponin/CK-MB assay. The capability to multiplex up to five analytes in a single assay has been demonstrated.
The company has developed a $15,000 laboratory analyzer and has placed about 15 systems for user evaluation. A handheld version of the analyzer has also been developed that costs about $3,000, but Natan believes the cost could drop to $1,000 in high volume.
Tracking down molecules and biomarkers . . .
Axela Biosensors (Toronto) has developed the dotLab system which uses Diffractive Optics Technology (DOT) for detection of biomolecules and microorganisms in a label-free format. The system uses a polystyrene test strip with antibodies or other specific binding elements attached to the surface in an ordered array. When target molecules or particles bind to the array, a diffraction grating is formed, allowing real-time detection and quantitation.
A prototype assay for the cardiac marker NT-proBNP has been developed with performance equivalent to that of existing immunoassay systems such as the Elecsys from Roche Diagnostics (Indianapolis), but which can potentially be performed at the point of care with imprecision of 10% or less. An enzymatic amplification technique has also been developed that provides enhanced sensitivity, down to 0.1 pg/mL for BNP. Axela has already commercialized the technology in the research market, with eight dotLab systems installed mainly in North America at about $50,000 each.
The company has licensed a microfluidics technology from Kimberly-Clark (Dallas) that is being used in combination with the DOT detection technology to develop a POC analyzer, dotKey/POC, for CLIA-waived testing. The analyzer consists of a laser and a photodiode detector, a simple configuration that lends itself to POC use. DOT is also a flexible modality that can be used to detect and quantitate particulate targets such as viruses and cells.
Singulex (St. Louis) is developing the Erenna Immunoassay System, which integrates a flow-through immunoassay format with single molecule detection to achieve high-sensitivity quantitative analysis of biochemical markers with a turnaround time of under 30 minutes. Singulex was the first company to demonstrate a cardiac Troponin assay with a sensitivity of 5 pg/mL.
Philippe Goix, PhD, said that one of the company’s goals is to employ its high-sensitivity assays to characterize normal levels of a wide range of disease markers such as troponin and IL-6 which normally are present at concentrations which are too low to be reliably measured with existing immunoassay techniques. The Singulex assays could then be used to screen patients for elevated levels of the target markers, potentially enabling earlier detection of serious diseases as well as tissue damage due to drug toxicity.
Disease screening tests being evaluated by Singulex include tests for early detection of cancer, diagnosis of Alzheimer’s, and early detection of stroke.
. . . and on to barcoding
Nanosphere (Northbrook, Illiniois) is commercializing a new labeling technology called Biobarcodes, along with its nanosphere particle labeling detection technology, with important applications in clinical diagnostics. Nanosphere versatility enables analysis of both nucleic acids and proteins, and the company has submitted two 510(k) marketing clearance applications for hemostasis assays. Additional assays are under development for measurement of cardiac Troponin I and prostate specific antigen, to be followed by an assay for cystic fibrosis screening.
Biobarcode technology enables very high sensitivity assays to be developed, opening up applications in early detection of conditions such as myocardial infarction, genetic disease, and cancer. In the case of cardiac troponin, Nanosphere is evaluating use of a high-sensitivity assay (with a 0.1 pg/mL detection limit) that may be able to detect cardiac ischemia at an early stage, before the condition progresses to a true myocardial infarction.
According to Greg Shipp of Nanosphere, who discussed the Biobarcodes technology at the conference, the troponin assay has allowed detection of patients who will eventually suffer a myocardial infarction well in advance of the event. That compares to existing troponin assays that do not allow detection of a myocardial infarction up to four-six hours after the event. The new Nanosphere assay is also being studied for applications in detection of cardiotoxicity in patients treated with various drugs, including pediatric patients.
POC for ‘low-resource’ settings
A key focus of this year’s meeting was POC testing and assay technologies for use in “low-resource” settings — formerly known as third-world countries.
POC testing now comprises about one-third of the clinical diagnostics market worldwide, with sales reaching about $11 billion in 2006 and continuing to grow more rapidly than the overall clinical diagnostics market.
Ove Ohman, PhD, of Amic (Uppsala, Sweden) described recent developments with the company’s Forecast technology, a chip-based microfluidic platform using the 4castchip, an injection-molded plastic device consisting of an array of micropillars which drive the flow of sample and reagent in a controllable manner. The underlying technology is based on fabrication processes and materials used in digital video disks.
The device can be used to perform assays on whole blood samples, and, mainly due to its highly controlled flow properties, provides precise results when used in POC settings. A proprietary coating technology is employed to reduce non-specific binding and facilitate attachment of antibodies to the surface.
Miniaturized, the device contains a number of active elements integrated into the design, such as a blood separation filter, a flow channel, a wicking zone and a unique detection zone using on-chip diffraction grating to more effectively couple light into the device and distribute emitted light to the detector.
POC assays have been developed for cardiac Troponin I (with a sensitivity of 0.03 pg/mL), TSH, CRP and IgE. The precision of the technology is exemplified by the CRP assay, for which the coefficient of variation is quoted at 6%.
The Forecast technology also has applications in the clinical lab, where microfluidics reduces the need for pumps and fluid handling devices.
Another platform employing microfluidics technology for use in POC testing — and in particular for infectious disease diagnostics in low resource settings — was described by William Rodriguez, MD, of Massachusetts General Hospital (Boston).
Public health disease-fighting
There is growing demand for diagnostic products for HIV/AIDS, malaria, and tuberculosis in low resource settings, driven in part by organizations such as the Clinton Foundation, which are funding disease-fighting efforts. Fewer than 10% of the 40.4 million people living with HIV infection worldwide have undergone an HIV test, and only 1% of these have access to the tests needed to manage their treatment.
The problem extends beyond public health to impact the economies of such countries, since in some cases projections indicate that up to 25% of the agricultural work force will be lost due to deaths from AIDS. This in turn has created a small but growing demand for easy-to-use, low-cost diagnostics, as shown in Table 1.
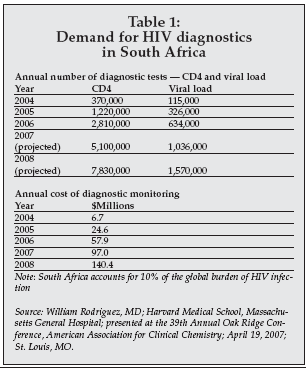 |
Rodriguez described microfluidic devices developed by his group that incorporate on-chip immuno-affinity chromatography for detection and can accurately perform CD4 cell counts using a 3 microliter whole blood sample in under two minutes, without the use of additional reagents, labels, or sample preparation. A similar approach is being used to develop microfluidic platforms for HIV viral load and tuberculosis testing.
Lumora (Cambridge, UK) is developing a low-cost platform for quantitative molecular diagnostics called Bioluminescent Assay in Real-Time (BART), with POC applications.
BART simplifies nucleic acid testing by using isothermal amplification, pyrophosphate chemistry and luminescence detection, combined with simple hardware, including a constant-temperature block and an optical detector. The method is capable of single-copy detection and is configured in a closed-tube format appropriate for non-laboratory settings. Applications under development include detection of Chlamydia in urine and sepsis detection.
BART also has been developed in a configuration suitable for the central lab, because offering low-cost, simple operation, rapid turnaround and quantitative real-time readout.
Gonzalo Domingo, PhD, of PATH (Seattle, Washington) discussed another microfluidic platform with applications in diagnostics for low-resource settings. PATH is developing three microfluidic disposable-based diagnostic platforms, including a PCR-based device for detection of bacterial pathogens in diarrhea; a device combining nucleic acid and immunoassay technology for detection of viruses and bacteria associated with measles, dengue, influenza, malaria, typhoid and Rickettsia; and a PCR-based platform for detection of sexually transmitted diseases and vaginal infections including Trichomonas vaginalis, Chlamydia, Neisseria gonorrheoeae, and bacterial markers of vaginosis.
The company’s Lab-on-a-Card device and portable reader, the DxBox, is a POC system that can perform an assay in 30 minutes with performance equivalent to central lab systems.
More POC technologies
Other new technologies for use in POC and low resource settings were described by speakers from Wave 80 Biosciences (San Francisco, CA) and Cleveland Biosensors (Brisbane, Australia).
As discussed by Daniel Laser, PhD, Wave 80 is developing a new micro-fabricated device platform with initial focus on malaria diagnosis, initially being developed for use by the U.S. military, and consequently has a rather high target cost of $10 per test. However, Laser said that the cost could be reduced 10-fold for high-volume applications in clinical diagnostics.
The device employs an electro-osmotic pump based on technology employed in semiconductor devices, a microarray-based reaction zone with immobilized antibodies specific for malarial pathogens (P. falciparum and P. vivax), and immuno-gold/immunofluorescence detection.
Test time with a prototype device is about three minutes using a 20uL fingerstick whole blood sample. A key advantage of the Wave 80 technology is the use of the high-pressure electro-osmotic pump, voiding the need to rely on capillary flow dynamics as in most other types of POC microfluidic devices.
Cedric Robillot, PhD, of Cleveland Biosensors, described the Biofiniti system, a handheld immuno-sensing device based on self-contained microfluidics, ferromagnetic actuation, magnetic capture, and electrochemical detection. The device uses a moving magnetic field coupled to a ferrofluid to pump sample and reagents through a network of microfluidic channels formed in an injection molded cartridge.
Assay sensitivity is enhanced via the use of liposome particles as labels. The liposomes contain an electroactive label that is released during the detection step by lysis, providing a high degree of signal amplification. The first assay to be developed is a test for microcystins, toxic peptides produced by cyanobacteria that can contaminate public water supplies.
The Cleveland Biosensors’ assay has a sensitivity of 0.1 nM, producing a result in 30 minutes compared to three hours typically required for existing methods. Another application under development is a rapid (10 minute) assay for the heart failure marker BNP, offering a sensitivity of 100 pg/mL, the typical threshold for BNP assays now on the market.
Cleveland Biosensors initially is targeting the food and water testing market, but the technology has obvious applications in clinical diagnostics. The company is collaborating with Biosite in its development program.
Nucleic acid sequencing
New technologies for molecular diagnostics were highlighted in various presentations at the Oak Ridge Conference. As shown in Table 2, the market for clinical molecular diagnostic products now exceeds $2.2 billion worldwide and is one of the most rapidly growing segments of the worldwide clinical diagnostics market. The focus at this year’s conference was, in particular, on nucleic acid sequencing technologies for diagnostics.
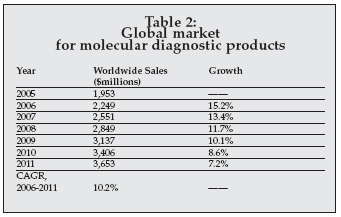 |
Determination of nucleic acid sequences is already an important application in molecular diagnostics, for analysis of viral genotype in patients with infectious diseases such as AIDS and hepatitis, as well as in genetic testing to detect mutations associated with drug response (pharmacogenetic testing) and genetic diseases. A variety of new technologies are under development for nucleic acid sequencing with potential applications in clinical diagnostics, as shown in Table 3.
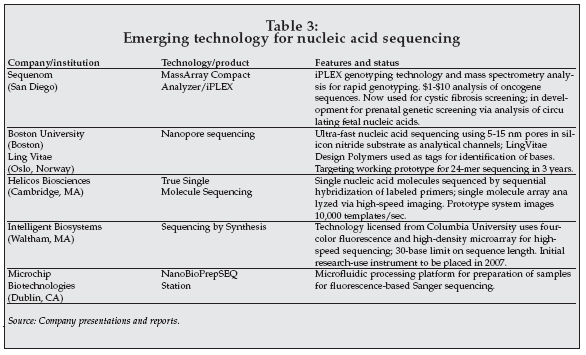 |
Charles Cantor, PhD, of Sequenom (San Diego), described the use of mass spectrometry in DNA sequencing, using the company’s MassArray system and associated iPLEX and MassEXTEND assay technologies. Mass spectrometry has so far been used mainly in protein analysis, as well for detection of toxic metals in blood samples. However, the technology is now showing promise in nucleic acid sequencing, where its ability to perform high precision measurements of molecular mass allows direct determination of a nucleic acid sequence.
Although the equipment needed for mass spectrometry is complex and expensive, fully automated systems are now available for sequencing which are suitable for the clinical lab setting. Cantor discussed applications in cystic fibrosis screening, screening for beta-thalassemia, detection of cancer-related mutations, genotyping of human papilloma virus, and quantification of SNPs in DNA extracted from maternal plasma for prenatal diagnosis of Trisomy 21 (Down’s Syndrome).
The versatility of mass spectrometric analysis is demonstrated by analysis of DNA methylation in patients with cancers such as acute myelogenous leukemia. Nevertheless, mass spectrometric sequencing requires use of complex and expensive equipment, and several hours are typically required for pre-processing of samples, pcr, and processing of samples prior to mass spectrometry analysis.
Nanopores, Sequencing by Synthesis, nanofluidics
Other new technologies for nucleic acid sequencing include nanopores, Sequencing by Synthesis and nanofluidics.
Amir Meller, PhD, of Boston University discussed development of nanopore array technology and its applications in sequencing. Nanopore arrays are being developed by Meller in partnership with LingVitae using that company’s Design Polymer technology.
Nanopores have been studied for a number of years as a potential vehicle for single-molecule analysis, including sequencing of nucleic acids. Nanopores can be formed using a variety of methods, such as by creating protein structures with naturally occurring channels having nanoscale dimensions, or by the use of a variety of fabrication techniques employed in the semiconductor industry such as e-beam cutting of nanopores in silicon nitride.
Meller is now using e-beam fabrication after finding that protein nanopores did not provide adequate discrimination of single bases. Miller is employing Design Polymer tags, which are 20-base nucleic acid polymers attached to each base in the sequence and providing added discrimination of bases, along with nanopore arrays to increase throughput relative to single nanopores. The goal is to reach a throughput of greater than 4 megabases per second using a 100x100 array, with a total cost for genome sequencing of under $1,000.
Helicos BioSciences is developing a new sequencing technology using single-molecule analysis. The assay involves immobilization of primers on the surface of a chip, followed by hybridization of sample genomic DNA fragments, and then by labeling one base at a time with a fluorescent tag.
An image of the chip surface is collected during each cycle, allowing the sequence of the target to be determined by monitoring the sequence of attachment of labeled bases. The method offers simple sample preparation methods, high throughput, and low cost. However, control of errors is an issue, necessitating the use of two passes to obtain an acceptable error rate of 0.01%.
The technology is being applied in oncology to guide treatment of lung cancer with agents such as Gefininib, which target specific oncogenes; and in personalized medicine to identify patients at risk for adverse drug reactions. Jerzy Olejnik, PhD, of Intelligent Bio-Systems (Waltham, Massachusetts), described his company’s program to commercialize Sequencing by Synthesis technology licensed from Columbia University (New York).
The technique is implemented in a high-density array on a chip having dimensions similar to a microscope slide. Four-color fluorescent labels are incorporated one base at a time, and fluorescent microscopy is used to determine the base binding pattern during analysis. The company’s goal is to develop a sequencer capable of a throughput of 3 gigabases per day at a cost of $1,500 per gigabase.
New dimensions in cell analysis
Cell analysis is another expanding area within clinical diagnostics, having moved beyond cell counting and flow cytometry to use of advanced automated imaging methods to determine cellular properties and assess response to drugs or other stimuli.
As shown in Table 4, products for cell analysis represent a significant and rapidly growing market. In addition to products for analysis of cellular specimens such as PAP smears, the field has advanced to include assessment of circulating tumor cells for prediction of cancer recurrence, analysis of cellular response to drugs or other stimuli for therapy guidance, and use of cells as sensors for sensitive detection of biological pathogens.
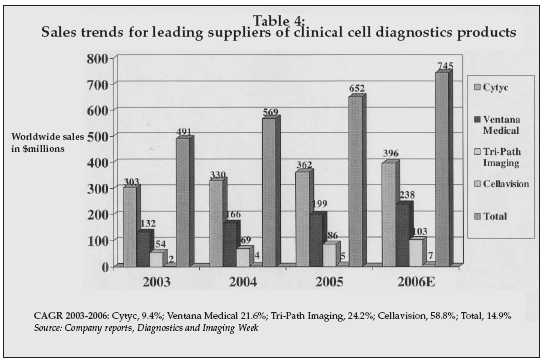 |
Advances in cell analysis for cancer were described by Garry Nolan, PhD, of Stanford University (Stanford, California). Nolan’s team has developed new multiparameter flow cytometry techniques that allow interrogation of cell signaling pathways in complex diseases such as Acute Myelogenous Leukemia, Follicular Lymphoma, and Systemic Lupus Erythematosus. Analysis of the flow cytometric data presents a new challenge due the very large amount of data generated in a typical analysis and the complexity of the signaling pathways.
The Stanford researchers developed a new electronic architecture for a statistics supercomputer which is used to analyze the data. The project is now focused on development of a comprehensive network topology map of signaling in all primary immune subsets, creating a generalized tool for assessment of a wide range of autoimmune disorders and malignancies. The technology may also have applications in monitoring of stem cell therapy. One near-term application involves assessment of B cells in SLE patients to rapidly predict response to drug therapy.
Douglas Malinowski, PhD, of BD Diagnostics Tripath Imaging (Durham, North Carolina), described new markers for applications in cell-based diagnostics for breast and cervical cancer.
While PAP smear screening has resulted in a significant reduction in the death rate from cervical cancer in the U.S., there is a 15%-25% incidence of false negatives, and current methods are unable to predict disease progression. There are similar issues with breast cancer screening using mammography and with prediction of relapse of breast cancer using lymph node analysis, and with the diagnosis of ovarian cancer.
Tripath has analyzed clinical specimens from patients with breast and cervical cancer to identify a panel of six protein biomarkers that can be used to improve detection and prediction of recurrence. Expression of any two or more markers indicates an increased risk for cancer or cancer recurrence.
Tripath has developed the VIAS Imager, an interactive histology imaging system, to perform quantitative analysis of protein biomarker expression patterns in cells, improving the detection of CIN2+ lesions within atypical cells in PAP specimens, and improving the predictive capability of tests used in early stage breast cancer.
Another cell imaging technology with applications in cancer diagnostics was described by Richard Bruce of Scripps-PARC Institute for Advanced Biomedical Research (Palo Alto, California). The Fiber Array Scan Technology (FAST) is used to detect rare circulating tumor cells that serve as an early indicator of recurrence in breast and lung cancer.
Tracking ‘circulating’ cells
A system for detection of circulating tumor cells in breast cancer patients developed by the Veridex unit of Johnson & Johnson (New Brunswick, NJ) has already received FDA clearance and is in clinical use. FAST technology promises to decrease analysis time, improve predictive value, and extend the applications of circulating tumor cell analysis to include diagnosis of lung cancer.
Because the FAST technology does not magnetically capture circulating cells, disruption of cell morphology doe not occur as with other systems. The analyzer employs a scanning laser beam to interrogate the cells, and a fiber optic detector to capture the emitted light. Positive cells are presented to the pathologist for verification following an automated scan. Initial studies indicate the VIAS system is more sensitive than the Veridex analyzer.
Two companies described development-stage systems that employ cells as sensors for sensitive detection of biological markers.
ACEA Biosciences (San Diego, California) described an integrated cell processing system, the BT-CES System, which measures changes in cell impedance to determine their response to various environmental stimuli using an electronic cell sensor array. The system has applications in cancer chemotherapy, immune system therapy, and infectious disease testing.
A drug resistance assay for use in predicting response to chemotherapy has been developed which measures cell impedance vs. time following exposure to a drug such as paclitaxel. The system has also been used to assess response to cancer immunotherapy using effector cells. In infectious disease diagnostics, the BT-CES system can be used to assess response to exposure to infectious agents such as West Nile Virus, and to assess the degree of protection provided by neutralizing antibodies.
Innovative Biosensors is developing the Cellular Analysis and Notification of Antigen Risks and Yields (CANARY) system for rapid detection of pathogens such as those involved in sexually transmitted diseases and respiratory diseases.
CANARY technology employs B cells engineered to express cell surface receptors that are specific for a pathogen of interest. Upon exposure to the pathogen and binding to the receptor, internal cell signaling pathways are activated resulting in stimulation of reactions that generate luminescence. The cells can be used to detect the presence of a specific pathogen rapidly, with a response time of one to three minutes depending on whether dry or liquid samples are tested.
A model system has been developed for detection of Chlamydia in urine with a sensitivity of 200 elementary bodies. CANARY requires minimal sample handling, making it ideal for applications in which rapid identification in non-laboratory settings is important.
