BBI Contributing Editor
ORLANDO, Florida — Molecular diagnostics has historically been used in the clinical lab for infectious disease diagnosis and screening, with those applications accounting for more than 95% of sales of molecular testing products used for in vitro diagnostic applications. As discussed by numerous presenters during the American Association for Clinical Chemistry (AACC; Washington) annual meeting held here this summer, however, the role of molecular diagnostics is now beginning to expand to include applications such as the diagnosis of hemostasis disorders, diagnosis of neurological disorders, cancer screening and diagnosis, early detection of diabetes, screening of newborns for genetic disorders (primarily cystic fibrosis), and selection and guidance of drug therapy (pharmacogenomics).
An example of an important new application in pharmacogenomics, and specifically in the guidance of preventative treatment for patients at high risk for cardiovascular disease, was described by Gualberto Ruano, MD, PhD, then-president of Genaissance Pharmaceuticals (New Haven, Connecticut), at an AACC plenary session. Genaissance is focused on the development of products for personalized medicine, and has embarked on a major effort to profile genes responsible for coding for receptors and enzymes, profiling genes for over 8,500 proteins since the company was founded five years ago. Of those, about 5,500 have been analyzed, resulting in the identification of over 140,000 SNPs and 130,000 haplotypes. An important finding is the discovery of universal rules that govern gene expression. The rules have been elucidated by analyzing the expression of multiple genes and promise to allow a major reduction in the complexity of genomic tests used in clinical applications. The research also has shown that the effect of gene and SNP variations on biological function of an enzyme or receptor produced by the gene depends on the affinity of the product, allowing researchers to better understand the role of a particular gene in causing or preventing a related disease.
The company and certain of its partners, which include Johnson & Johnson (New Brunswick, New Jersey), Pfizer (New York), Biogen (Cambridge, Massachusetts) and AstraZeneca (London), are sponsoring the STRENGTH trial to study the use of haplotype expression analysis in predicting response to statin therapy. The drugs studied include Pfizer's Lipitor, Pravachol from Bristol-Myers Squibb (Princeton, New Jersey) and Zocor from Merck & Co. (Whitehouse Station, New Jersey). The trial is studying 175 candidate genes in 150 patients at both the lowest and highest recommended dose. The initial results of the trial have shown a 45% overall average drop in LDL levels as a result of statin treatment, but there are considerable differences in response between patients, presumably due to differences in genetic characteristics. Genetic markers have been identified that allow categorization of patients based on their response to statin treatment, in principle allowing a patient to be tested prior to starting therapy to determine which drug should be used to obtain the best response. Additional markers have been found that indicate if a high or low dose of the drug should be used. An estimated 36.5 million people in the U.S. are eligible for treatment with statin drugs based on their risk profile.
Another set of markers is being studied by Genaissance in the CARING study, which is evaluating the use of pharmacogenomic testing to identify the 10% of epilepsy patients who have an adverse response to clozapine. In addition, pharmacogenomic assays based on markers discovered by Genaissance have been developed for the ProbeTec ET system manufactured by BD (Franklin Lakes, New Jersey) to assess asthma patients for their response to albuterol therapy. The asthma tests are based on a set of 14 SNP markers discovered by Genaissance that predict drug response. However, fundamental rules governing gene expression discovered by Genaissance allow response to be determined by measuring only six markers, significantly reducing the complexity of the assay. The expression rules apply to all gene expression, and thus should allow a similar reduction in the number of SNP markers required in all types of clinical pharmacogenomic assays. The ProbeTec assay allows 96 genotypes to be analyzed in about 75 minutes, and can be performed using urine, bucchal swabs, or blood specimens.
Ruano said he envisions the development of a genetic Physicians' Desk Reference that can be used to guide drug selection and dosage based on a patient's DNA characteristics. The number of tests required for selection of a drug to treat a given disease will be reduced to tens, not hundreds or thousands as might be expected based on the number of SNP markers, by the use of the expression rules discovered by Genaissance. The initial clinical applications of the technology are likely to be in the treatment of patients with highly complex conditions, such as diabetics with co-morbidities, where pharmacogenomic tests will provide guidance to avoid the complex empirical process of therapy selection used at present. Genaissance has not yet announced plans for commercialization of its pharmacogenomic assays.
Another strategy for creating personalized approaches to disease management is to use proteomic, rather than genomic, markers to determine disease predisposition and to select an appropriate therapeutic agent. Since proteins are the molecules that carry out biochemical functions in the body and are often modified after being produced by genes, they are typically the preferred targets for drugs, rather than the genes themselves. As discussed by James Witliff, PhD, of the University of Louisville (Louisville, Kentucky) at the AACC conference, at least seven companies are now developing proteomic microarrays that are expected to have applications in clinical diagnostics. Witliff is using microarrays manufactured by the leader in the field, Ciphergen Biosystems (Fremont, California), along with Surface Enhanced Laser Desorption Ionization (SELDI) read-out technology in studies aimed at identifying new cancer markers. The studies use cell extracts or plasma from tissues isolated from human tumors using a technique called Laser Capture Microdissection from Arcturus Engineering (Mountain View, California) that allows only known tumor cells to be segregated from a tissue specimen. Although the system is not yet available for clinical use, costs have dropped to a level of about $9 per well, with chips consisting of eight or 16 wells per chip. Witliff's studies are aimed at identifying markers that can be used as chemotherapy drug targets, as well as to characterize an individual patient's tumor for personalized therapy.
Myriad Proteomics (Salt Lake City, Utah) is another leading company in the proteomics field. Myriad's initial strategy was to create a comprehensive map of the entire human proteome, but the company has now adopted a more focused approach that targets the identification of specific proteins that can be used as drug targets. Myriad's search strategy uses a three-tiered approach that includes assessment of protein distributions in normal vs. diseased tissues such as tumors; analysis of protein structures, and evaluation of protein-protein interactions to identify molecules that play important roles in disease processes and that will provide effective drug targets. There are an estimated 120,000 different proteins coded for by the about 40,000 known genes, and only about 5,000 of those proteins have been characterized. Myriad, which has 80 employees, is also taking advantage of data from its pharmaceutical partners (which include 10 major drug companies) to help direct its search at the most promising and commercially attractive drug targets.
Nanogen (San Diego, California) has made significant progress in developing microarray devices for clinical applications. The company has introduced a genomic microarray for cystic fibrosis (CF) screening, an application that is rapidly developing into the first truly high-volume clinical molecular genetic assay. The Nanogen assay detects the 25 most prevalent CF mutations, which occur at rates of 0.1% or higher in the population. The disease afflicts about one in 29 Caucasians, but is less prevalent in the Asian population. Guidelines published by the American College of Medical Genetics (Rockville, Maryland) and the American College of Obstetricians and Gynecologists (Washington) in March 2001 recommended that CF screening be offered to all non-Jewish Caucasian and Ashkenazi Jewish couples planning a pregnancy and to those seeking prenatal care, and that other ethnic groups be made aware of the risks of the disease and of the availability of screening tests. According to Wayne Grody, MD, PhD, of UCLA School of Medicine (Los Angeles, California), who discussed the topic at the AACC conference, between 57% and 97% of carriers are detected by the screening process.
The Nanogen test, which will be available this fall, will use analyte-specific reagents (ASRs), avoiding the necessity to obtain full FDA approval as is required for most test kits. The microarray test cartridges employed for the assay can screen 15 samples simultaneously, and the NanoChip Molecular Biology Workstation used to analyze the arrays is capable of processing 120 samples per day in two batches of 60 each. Six markers are analyzed in the initial screening run for each patient, including the delta F508 mutation, and individuals who test positive for markers other than delta F508 (about one in 100) are analyzed for additional markers. About two hours of hands-on time is required to run 60 samples. Other tests developed for use on the Nanogen system include SNP marker panels for Factor V Leiden and the PT20210 mutation that confers increased risk for deep venous thrombosis; and a NAT2 genotyping assay that characterizes 13 SNP markers that indicate abnormally low metabolism of drugs used in the treatment of infections (isoniazid and sulfonamides) as well as of carcinogens.
Roche Diagnostics (Indianapolis, Indiana) is another supplier of molecular diagnostic products with applications in pharmacogenomic testing, as well as for a number of infectious disease testing applications. Roche already has introduced an ASR chip designed by Affymetrix (Santa Clara, California) that tests for cystic fibrosis mutations, and previewed the new Cobas TaqMan 48 real-time PCR system at the AACC exhibition, with launch scheduled at the end of 2002. ASR reagents for performing Hepatitis B and C analysis will be available for the new system. Because it is designed to be an open platform, the TaqMan 48 also will allow users to perform other types of molecular tests.
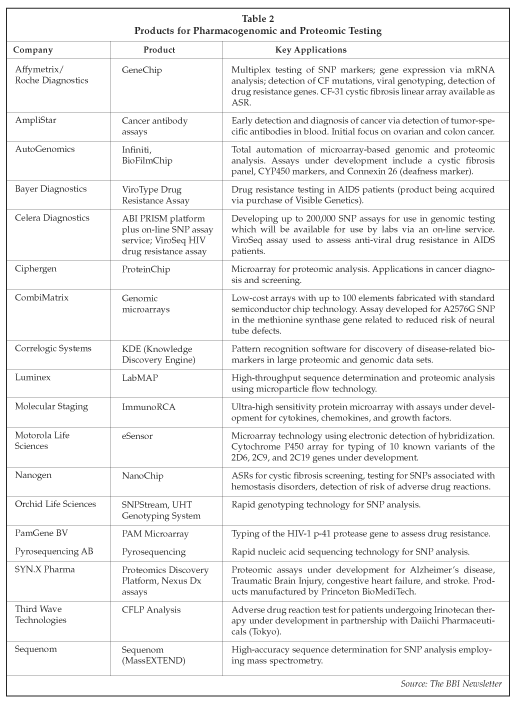
Other suppliers of pharmacogenomic and proteomic products with applications in clinical testing are listed in Table 2, along with their products and key applications. In addition to those listed in the table, other products with applications in the preparation of samples for genomic and molecular diagnostic testing include the BioRobot 9604 and the new MDX from Qiagen (Hilden, Germany), the newly launched Viper from BD, the Procleix system from Gen-Probe (San Diego, California), the NucliSens Extractor from bioMerieux (Marcy l'Etoile, France), the MagNA PURE LC from Roche, an automated nucleic acid extraction system from AutoGen (Framingham, Massachusetts) and the Arteas LabCard from Aclara Biosciences (Mountain View, California).
 |
AutoGenomics debuted its Infiniti system for the first time at the AACC exhibition. The system features unique BioFilmChip technology and combines all processing and analysis steps in a single, integrated system. A wide range of applications are under development, including a cystic fibrosis panel that detects up to 31 mutations at a cost of from $50 to $450 depending on the number of markers analyzed. Both hybridization and FISH assays can be accommodated on the platform, as well as proteomic assays. Up to 24 different microarrays can be loaded on the system at one time in two magazines that hold 12 chips each. The system is currently undergoing clinical trials, with product launch scheduled for 4Q02. The system will be the first fully integrated and automated molecular diagnostics analyzer on the market, according to AutoGenomics. Initial applications will focus on women's health, newborn screening, cardiovascular disease management and cancer screening.
Another important application for genomic and proteomic testing is cancer screening and diagnosis. A number of research groups are investigating the potential to use panels of SNP markers or expressed proteins to identify cancer at an earlier stage, and to assist in making a more detailed diagnosis that will allow improved selection of chemotherapeutic agents. As discussed by Lance Liotta, MD, PhD, of the National Institutes of Health (Bethesda, Maryland) at the AACC sessions, technology is now available that allows a panel of proteomic markers to be analyzed to characterize an individual patient's cancer. A drug cocktail can then be selected that will attack multiple nodes of the tumor's biochemical network simultaneously, avoiding the escape phenomenon that typically occurs in patients undergoing chemotherapy. The approach also will allow lower doses of each agent to be used since the drugs will target specific effectors, reducing the adverse side effects of chemotherapy. Liotta is using the ProteinChip from Ciphergen to analyze up to 300 specimens per day, focusing on the identification of new markers for prostate and ovarian cancer. Tests are planned that can be performed by a CLIA-certified reference lab established by Liotta at the NIH as part of an ongoing clinical trial aimed at discovering markers that allow earlier detection of cancer recurrence and improved guidance of alternative therapy selection. Cancers of specific interest include prostate, breast, and ovarian cancer. A recent study of the use of proteomic markers for the early detection of ovarian cancer conducted by Liotta and his colleagues found that a sensitivity of 99% and a specificity of 95.5% could be achieved, but that level of performance is not yet adequate for use in general population screening due to the high number of false positives that would be expected.
One potential issue for developers of microarray devices for use in clinical pharmacogenomic and proteomic testing is the changing stance of the FDA regarding regulatory status for the products. As discussed by Steve Gutman, MD, of the FDA, pharmacogenomics presents unique regulatory challenges, and the agency is continuing to assess the technology to determine how it should be handled from a regulatory perspective. Most importantly, Gutman said that the new philosophy within the agency is that such products should be evaluated on an ongoing basis, not just during the approval phase, and that companies developing microarray-based ASR products in particular may be in a risky position, with a possibility that the regulatory requirements may be changed in the future. Clinical labs tend to favor the use of FDA-cleared kits rather than ASRs, since that approach typically saves time and money and provides more standardization.
