Items Tagged with 'Arc Institute'
ARTICLES
Drug design, drug delivery & technologies
Epigenetic switch and gene editing activate human T cells
Read MoreDrug design, drug delivery & technologies
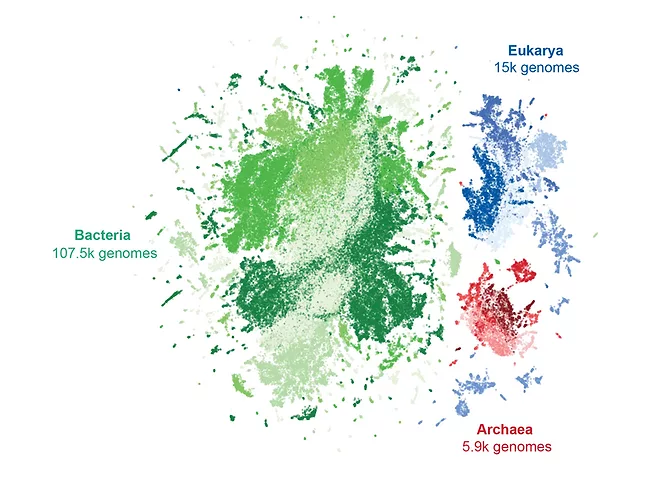
Evo 2 AI allows genome and epigenome modeling of all life domains
Read MoreDrug design, drug delivery & technologies