BBI Contributing Editor
SAN FRANCISCO Molecular diagnostics continues to be the most rapidly growing segment of the clinical diagnostics market, and has now become the fifth largest segment of the global diagnostics market. Although both the unit and dollar volume molecular diagnostics market remains dominated by infectious disease testing products, other segments are rapidly gaining in importance, including genetic testing, cancer diagnostics, testing for bioterrorism agents, and pharmacogenetic testing. The 19th annual San Diego Conference, sponsored by the American Association for Clinical Chemistry (AACC; Washington), and held here in mid- November, provided a window on new developments in molecular diagnostics technology that promise to enable further expansion in the range of applications and to increase the level of automation of molecular testing.
Lab-on-a-chip technologies are beginning to show promise for molecular testing, and bioinformatics is playing an important enabling role in certain new applications. Advanced detection technologies, continue to improve, and techniques to enhance the quality of microarray test data are under development that will be important in enabling such devices to be widely used in the clinical lab.
Advances in detection technologies
One promising technology now being applied to enhance detection capabilities in molecular assays is quantum dots. Quantum dots are small-diameter (3 nanometer) particles that can be coupled to molecules used as reagents in molecular assays to provide highly sensitive detection as well as positional information. As discussed at the San Diego Conference by Sanford Simon, PhD, director of the Laboratory for Cellular Biophysics at Rockefeller University (New York), quantum dots can be used to track molecular events such as transfer of proteins through membranes. Because of their unique ability to provide total internal reflection of light, the particles can provide spatial resolution as low as 100 nm, a level that cannot be achieved by optical microscopy. Simon has demonstrated the ability to track liposomes fusing with cell membranes, and to track changes in such processes due to exposure to various agents. Quantum dots are roughly the same size as some molecules used in conventional immunoassay labeling and do not interfere with reactions or cell function. In addition to uses in in vitro cellular assays, quantum dots can be used to track cells in vivo, as demonstrated by Simon in experiments in which tumor cells infused into animals have been tracked to determine where they become localized. In the clinical lab, quantum dots have additional applications in allowing rapid detection of multiple analytes, since the particles can be manufactured to emit fluorescence at a wide variety of different wavelengths.
Another new detection technology with applications in clinical molecular diagnostics is QUAL probes. QUAL probes, as described by Eric Kool, PhD, of Stanford University (Palo Alto, California) at the conference, are nucleic acid probes that emit fluorescence only when hybridization occurs. As a result, there is no need to wash away unbound labeled probes, greatly simplifying the assay format. As compared to molecular beacons, which are another type of highly sensitive fluorescent label with applications in molecular diagnostics, QUAL probes exhibit higher sensitivity, due in part to a signal amplification technique developed by Kool that uses universal flexible linker probes to partly de-stabilize probe hybridization. In addition, Kool has recently incorporated fluorescence resonance energy transfer labels into the QUAL probes to reduce background and further improve sensitivity. However, amplified QUAL probes require a somewhat longer detection time than molecular beacons due to the need for incubation. Kool is developing applications of QUAL probes in oncogene detection, identification of pathogens and their drug resistance genes, and in vivo imaging of tumors in the lungs and skin, as well as rapid genotyping of leukemias. The technique may allow leukemias to be rapidly genotyped directly from a blood specimen using a simple fluorescence microscopy test, rather than a complex flow cytometry assay.
Microarrays represent another detection technology that is playing an increasingly important role in clinical diagnostics. Affymetrix (Santa Clara, California), the leading supplier of microarrays, reported 2003 sales of $281 million, and sales have grown at a 26% compound annual rate (based on projected 2004 sales of $316 million) over the past five years. As discussed by Richard Rava, PhD, the company's senior vice president for research, newly emerging concepts about the mechanisms of gene expression indicate that microarrays may play a key role in analyzing the complex expression pathways involved in both normal processes as well as in disease states. As discussed by Rava, the original paradigm that one gene creates one protein does not hold true in biological systems. Instead, as a result of alternative RNA splicing, more than 60% of human genes produce more than one product. At present, only about 50% of the resulting gene transcripts produced by the human genome have been characterized. Affymetrix has developed a new, lower-cost gene chip scanning system employing dual interacting laser beams and a CCD detector to rapidly analyze a large number of probes on a single microarray chip. Based on a feature size of one micron in the current Affymetrix arrays, and the ability to use 15-mer probes at each location on an array, about 1 billion different probes can be incorporated on one chip. The use of dual lasers allows patterned excitation to be performed, effectively giving a readout resolution that is less than that which can be achieved with conventional optical scanning. Although the existing Affymetrix arrays are expensive, costing $3,000 or more per chip for research-use devices, Rava said he believes whole genome scanning chips can be developed that can analyze 10,000 sequences that will be inexpensive. About 3 million chips of that type can be produced from a single wafer.
Another new development that promises to allow lower cost tests to be produced is high-resolution DNA melting analysis, described by Carl Wittwer, PhD, of the department of pathology at the University of Utah (Salt Lake City). Although melting analysis has historically been considered a low-resolution method, Wittwer has developed techniques for rapid, high-resolution analysis that can be used in conjunction with real-time polymerase chain reaction (PCR) to provide high throughput, cost-effective genotyping. An assay for Factor V Leiden mutations using DNA melting analysis has already received FDA clearance. Idaho Technology (also Salt Lake City) is commercializing the technology in the form of a 96- or 384-well format LightScanner instrument, to be launched in 2005. Key advantages of melting analysis include elimination of the need to develop probe sets for detection of specific mutations, closed tube analysis and the ability to provide genotype data for multiple sequences in a single analysis. Wittwer described a cystic fibrosis (CF) screening assay based on melting analysis that allows detection and genotyping of CF mutations. Wittwer said he believes that up to 99% of all DNA sequencing applications can be addressed more rapidly and cost-effectively with DNA melting assays.
Xing Su, PhD, of the biotechnology research group at Intel (Santa Clara, California), described new technologies under development at the leading microelectronics company that may have important applications in molecular diagnostics. Intel has the capability to manufacture microelectronic devices with a feature size that is similar to that of biological entities. The company is pursuing research aimed at combining its capabilities in nanotechnology with biotechnology to create new devices with applications in fields such as molecular diagnostics. Su described a new labeling and detection technology called Composite Organic-Inorganic Nanoparticles (COINS), comprised of silver nanoparticles glued together by organic labels that produce Raman scattering of incident light. COINS can be produced to emit multiple wavelengths by using combinations of organic molecules, allowing the particles to have different Raman signatures, and thereby enabling multiplexed analysis. By combining COINS with microarrays, which also provide multiplexing capability, a system can be created that offers a very high degree of multiplex analysis as well as extremely high sensitivity, potentially allowing single molecules to be detected. Model systems have been studied that show a sensitivity that is 10,000-fold better than ELISA, as well as a dynamic range of 105 vs. 103 for ELISA. Su has demonstrated that COINS can be used as labels in immunoassays, and applications to other types of molecular assays appear feasible. Su's team is collaborating with the Fred Hutchinson Cancer Center (Seattle) in the development of clinical applications for the technology.
Scott Eastman, PhD, of Quantum Dot (Colorado Springs, Colorado), described his work with the company's nanocrystal labels combined with paramagnetic beads to perform sensitive multiplexed nucleic acid assays of gene expression. Quantum dots can be synthesized to provide up to 300 spectral color codes to allow the expression of multiple genes to be analyzed in each well of a 384-well microplate. Readout can be performed with high sensitivity to detect multiple targets in each well with a precision of about 5%. About 1 million copies of a target nucleic acid sequence can be detected without resorting to amplification. Using T7 amplification, a sensitivity of 10,000 copies has been achieved. Eastman has demonstrated the capability to assess differences in gene expression between tissues such as liver and pancreas, and can detect a difference of as little as 1.4-fold in expression. The technology compares favorably to quantitative PCR in terms of sensitivity and throughput, according to Eastman, and is now being applied to SNP detection as well as analysis of variants arising due to alternative splicing.
Douglas Lane, president of ViaLogy (Altadena, California), described his company's signal processing software and its applications to DNA microarray analysis. The company has developed a technique called Quantum Resonance Interferometry (QRI) that allows the detection of ultra-weak signals, even those below the noise background. The technique involves modeling of the noise background, and detection of the disturbance in the background due to a signal. In analysis of data from Affymetrix microarrays, ViaLogy has demonstrated a 50% improvement in precision, as well as the ability to detect one to two gene copies per cell. Lane said he believes it may be possible to analyze gene expression from a specimen consisting of only a few hundred cells with the use of PCR. That capability may allow the use of fine needle aspirates to perform a comprehensive gene expression analysis of a tumor, or to identify stem cells in bone marrow aspirates for use in cell therapy. ViaLogy is collaborating with IBM Life Sciences (White Plains, New York) and Agilent Technologies (Palo Alto, California) to assess uses of QRI in biological applications.
LOC, integrated diagnostics technologies
In addition to advances in detection technologies, a number of emerging technologies were described at the San Diego Conference that may allow development of fully integrated microanalytical systems for use in molecular diagnostics. An overview of the technologies discussed at the meeting is shown in Table 1. Richard Mathies, PhD, of the University of California-Berkeley described research with microcapillary arrays that have applications in rapid genotyping using nanoliter to subnanoliter sample and reagent volumes. The arrays are fabricated in a radial configuration, and include integrated nanoliter valves, on-chip electrophoresis channels and a polymer capture gel for purification, heated chambers for performing pcr reactions and microbeads with primers attached for performing sequencing using the Sanger reaction, all on a single chip. Mathies is collaborating with Microchip Biotechnologies (Fremont, California), which is planning to commercialize the technology. Initial efforts are focused on development of a portable device for infectious disease testing, and assays for E. coli 157:H7 and E. coli 055:B5, as well as for methicillin-resistant staphylococcus aureus, have been prototyped which offer a 30-minute turnaround time. Another application under development is forensic testing with a portable system configured as an integrated microfluidic short terminal repeat analyzer (GATTACA-TO-GO). Microchip Biotechnologies and Mathies are collaborating with the Division of Forensic Science of the State of Virginia and with the Palm Beach, Florida, Sheriff's Office to implement the technology for forensic applications. A rapid PCR assay for sex determination using a bucchal swab sample has been developed that involves collecting the sample with a swab, placing the swab into a hot PCR mix, and then applying the resulting mixture to the chip analyzer. It also may be possible to use blood samples in the analyzer, according to Mathies, although issues arise due to interference by heme proteins with the PCR reaction. One solution may be to collect the blood samples with microneedles to eliminate red blood cells as part of the sampling process.
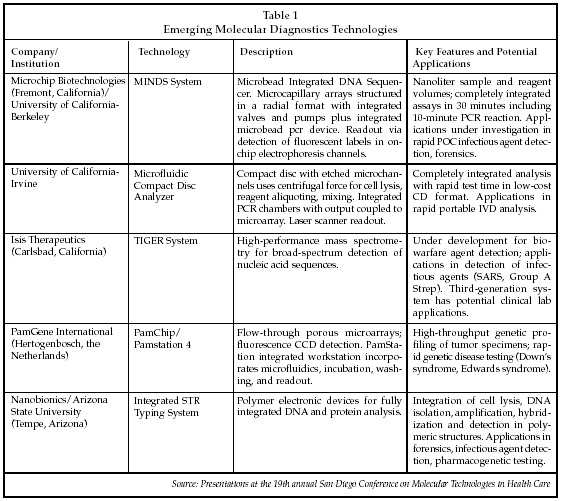
Another new approach to integrated microfluidic molecular diagnostics using a compact disc as the analytical platform was described by Marc Madou, PhD, of the University of California-Irvine. As discussed by Madou, the technology embodied in a standard optical disc drive is actually a high-frequency (1 MHz) scanning laser microscope. By combining a CD drive with the centrifugal forces created by spinning of the disk, he has succeeded in creating an integrated sample processor/microarray analyzer. In one configuration, glass slides are incorporated into the plastic disc and microcapillary channels are formed by etching followed by overlay of a polymeric membrane. Samples are injected near the center of the disk, and the disk is then rotated at 450 rpm to force the specimen outward through a series of processing and mixing steps, including cell lysis, sample splitting, molecular separation, PCR, DNA hybridization and microarray detection using a scanning laser/detector device. Madou has demonstrated the ability to perform DNA hybridization in two to three minutes. The CD technology is particularly well suited for in vitro diagnostics applications because it minimizes the number of different materials that contact the sample and the reaction mixture, avoids the need for chemical lysing reagents, and minimizes the influence of sample composition on the microfluidic processes.
Rinie van Beuningen, vice president of technology for PamGene International (Hertogenbosch, the Netherlands), described his company's programs to develop new integrated molecular analysis systems using proprietary porous microarrays. PamGene's goal is to develop an FDA-approved platform capable of detecting 10 to 400 genetic markers using a single microarray with a one-day turnaround time. PamGene was spun off from Organon Teknika/Akzo Nobel (Arnhem, the Netherlands) in December 1999 to commercialize a proprietary microarray technology. Licensees of the PamGene technology include Olympus (Tokyo) in the research market and Innogenetics (Ghent, Belgium) in the diagnostics market. The company is venture capital-funded, having raised EUR 16.3 million over the past four years.
The PamGene microarrays are comprised of porous ceramic material and microfabricated using a low-cost, high-volume manufacturing process. Various chemical reactions can be carried out by using low-level pressure to force liquid reagents into the chip, and then reversing the pressure to extract the reagents prior to the next processing step. PamGene has demonstrated the capability to perform hybridization reactions in minutes, as well as the ability to precisely quantitate reaction products with a coefficient of variation of 6%. Various readout methods can be employed, including fluorescence labeling and DNA melting analysis. Applications under development by PamGene include genetic screening and related pre-/post-natal screening tests, diagnosis/risk assessment for colorectal and breast cancer, high-resolution HLA profiling, and molecular karyotyping. A post-natal genetic screen can be performed in about 5.5 hours starting with a blood sample, and an initial blinded pilot study found an accuracy of 100% in detecting various trisomy syndromes.
Nanobionics, a company founded by Frederic Zenhausern, director of the Center for Applied NanoBioscience at Arizona State University (Tempe, Arizona), is developing a new integrated microanalytical platform based on polymer electronics. Polymeric electronic materials already are being developed for applications in portable imaging devices and handheld computers. Zenhausern discussed uses of the technology for low-cost molecular diagnostic devices using in-line manufacturing of polymeric cartridges that provide integration of cell isolation, cell lysis, DNA isolation, amplification, hybridization and detection via an on-chip near-field optical detector using surface-enhanced Raman techniques. The fabrication process relies on proven technology adapted from the semiconductor industry. Various electrochemical techniques have been developed for sample manipulation, including cell lysis and pre-concentration, separation of reaction products, molecular stretching of DNA, and molecular trapping. An integrated STR typing system is being developed under a contract with the Federal Bureau of Investigation (Washington) that consists of a 20 cm x 30 cm analyzer using polymer cartridges. In addition, Zenhausern is collaborating with the Mayo Clinic (Rochester, Minnesota), the Scottsdale Health Center (Scottsdale, Arizona) and the University of Arizona Cancer Center (Tucson, Arizona) to develop polymer electronics for clinical diagnostic applications.
Analysis of the data generated by integrated microanalytical platforms to derive clinically useful information presents another challenge to implementation of such technology in the diagnostics market. GeneGo (St. Joseph, Michigan) is developing bioinformatics technology to address that challenge using a platform called Metacore. Metacore is an Oracle-based database comprised of 150 relational tables that describe the biochemical pathways associated with expression of greater than 90% of known human genes. The Metacore platform can depict the pathways of protein-protein interaction, protein-DNA interaction, and protein-drug interaction, and the database currently includes about 2 billion five-step pathways derived from the scientific literature. GeneGo is developing algorithms to analyze biological networks that can be used to, for example, predict the effects of new drugs. A dedicated platform, MetaDrug, has been developed for analyzing drug response data. In the clinical diagnostics arena, applications include assessment of the differences in response to various therapies for glaucoma and breast cancer, as well as identification of non-invasive vs. invasive vs. late-stage cancer based on an analysis of protein expression patterns. Specific applications have been developed for use in analyzing data generated by Affymetrix and Agilent microarrays. The GeneGo software can be accessed via the web or by installing an in-house server running the MetaCore database.
Cerner Medical Informatics (Kansas City, Missouri) has developed a new bioinformatics system called PathNet Helix that can be used to analyze genetic test data obtained from molecular diagnostic testing devices used in the molecular pathology laboratory. The first installation of PathNet Helix was scheduled to go live in December. PathNet Helix automatically captures genetic test data through Cerner's Millennium open clinical information system, making it a continuously evolving database that can be used to analyze patient test results using the latest genomic research findings. The system provides genome-based reference data that can be searched to extract interpretative information pertaining to a test result, and can also support comparative analysis of data obtained on other patients to identify common mutations or to detect genetic changes occurring in a particular patient over time via the analysis of serial test data. For example, it can allow tracking of the evolution of gene defects in a cancer patient. In addition, the system can provide alerts to physicians when a specified course of therapy is inconsistent with the patient's genetic characteristics. If an anti-HIV drug is ordered for a patient with a documented resistance mutation, the system will raise a flag for the physician. Cerner updates the PathNet database monthly based on input from its scientific advisory board and new data collected by the system through the LIS network.
Another important topic discussed at the San Diego Conference that plays a key role in ensuring that molecular diagnostic testing benefits patients is quality assurance strategies for genotyping tests, including strategies for quality assurance of microarray data. Gene Express (Toledo, Ohio) has developed a line of DNA standardization assays, called StaRT-PCR, which provides standardized numerical gene expression values, or Standardized Expression Measurement. The StaRT-PCR products provide laboratories with a tool to meet the FDA's requirements for multigene expression assay methods which stipulate a sensitivity of less than 10 molecules, a day-to-day reproducibility of better than 15%, 5% to 10% intra-sample variance, and the ability to detect a 20% difference in expression. Those requirements were implemented not for clinical diagnostic applications, but to allow the agency to deal with the need for standardization of microarray data being submitted by pharmaceutical manufacturers as part of their clinical trial documentation. The product includes a standardized mixture of internal standards (SIMS) that is analyzed as part of each expression assay. For clinical diagnostics, the StaRT-PCR products have potential applications in improving the diagnosis of lung cancer vs. conventional morphological analysis by allowing quantitative comparisons of normal vs. tumor tissue gene expression to be performed, as well as improving the ability of gene expression assays to predict response to chemotherapy.
New applications attract clinicians' interest
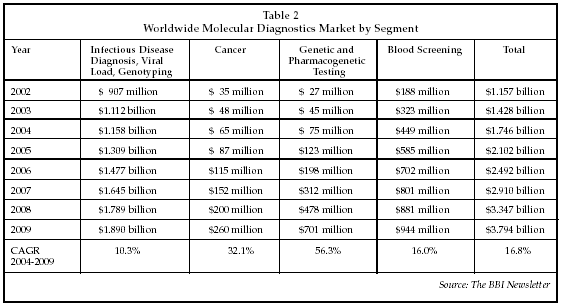
The expanded array of technologies applicable to molecular diagnostics, as well as advances in disease-related gene discovery and improvements in bioinformatics technology for analyzing test data, have engendered growth in the number of applications of molecular diagnostics in the clinical lab. Although the molecular diagnostics market was initially limited almost entirely to applications in infectious disease testing, there recently has been a rapid expansion in the uptake of new applications in the areas of genetic testing, cancer diagnosis, and pharmacogenetics. As discussed at the conference by Deborah Payne, PhD, director of the molecular diagnostics laboratory at the University of Texas Medical Branch (Galveston, Texas), there has been a dramatic increase in physician orders for new molecular tests within the past two years. Payne's laboratory serves a geographic area that covers about one-third of Texas, and performs an average of 1,800 tests per month. Although infectious disease tests still comprise the majority of the total, the number of genetic tests per month has increased by 600% since the beginning of 2004, and the number of pharmacogenetic tests has increased 100% to 75 per month. In addition, the number of solid tumor tests has increased 300% over the past two years, to about 300 per month. As shown in Table 2 below, growth rates in the global market for molecular diagnostic testing products reflect similar trends, with projected growth rates for cancer and genetic/ pharmacogenetic products far outpacing the growth in the infectious disease segment. The global market is expected to exceed $1.7 billion in 2004, and sales will approach $3.8 billion by 2009.
Additional new applications of molecular diagnostics were discussed at the San Diego Conference by Ronald Davis, PhD, director of the Stanford Genome Technology Center (Palo Alto, California). One application involves microarray analysis of RNA in trauma patients to improve the ability to determine a prognosis. At present, according to Davis, there is not a good correlation between clinical symptoms and data from clinical exams performed on trauma patients with outcome. In collaboration with Ingenuity Systems (Mountain View, California), which provides a systems biology database incorporating knowledge derived from genome studies, and ParAllele (South San Francisco, California), developer of a microarray that can be used to interrogate multiple SNP changes, Davis is analyzing protein and gene expression in trauma patients to detect patterns that may serve as prognostic indicators. A microarray with 300,000 features is to be evaluated for broad-spectrum analysis of changes in expression in trauma. The goal of the program is to develop a hand-held, battery-powered instrument for use in assessing patient prognosis in the trauma setting.
Kathleen Danenberg, president and CEO of Response Genetics (Los Angeles), discussed her company's collaboration with Roche Diagnostics (Indianapolis) to develop predictive tests for chemotherapy response. The goal is to identify genetic markers that can provide guidance for all patients undergoing chemotherapy. Progress has been made in identifying useful markers, as shown by gene expression studies in patients treated with 5-fluorouracil. The use of expression analysis of thymidylate synthase has been studied extensively, and shown to provide some predictive power, but the addition of dihydropyrimidine dehydrogenase expression increases the predictive power to 90%, and if thymidine phosphorylase is added a 100% predictive power has been demonstrated. While the results of initial studies are promising, clinical adoption of such testing has been hampered by the lack of availability of fresh frozen tissue specimens for use in large-scale studies. Until recently, gene expression assays performed on formalin fixed paraffin-embedded (FFPE) specimens, which are the primary specimens available for clinical studies, were highly imprecise. However, Response Genetics has developed new approaches for FFPE tissue analysis that have significantly improved the ability to isolate large quantities of non-degraded nucleic acids, achieving about a ten-fold higher yield than previous techniques.
The company has now analyzed about 18,000 tissue specimens for gene expression in cancer patients, and can obtain gene expression data the same day the specimen is collected. The process also relies on microdissection of the tissue specimen to ensure that tumor rather than normal tissue is analyzed. Danenberg said she believes the use of gene expression analysis to guide chemotherapy is about to undergo a major expansion in utilization. Related applications under development by Response Genetics include selection of responders to Gemcitabine, a drug used in the treatment of lung cancer; and identification of patients with high expression of epidermal growth factor receptor (EGFR), who are likely to respond to drugs such as Iressa. Studies also are under way to analyze gene expression in lung, colon and breast cancer, using the Affymetrix X3P chip for analysis. Response Genetics has partnered with Arcturus Bioscience (Mountain View, California), a manufacturer of Laser Capture Microdissection instruments, to market a line of reagents called Paradise used in tissue gene expression analysis. Arcturus also is pursuing development of tests using FFPE samples to improve the ability to identify the source of a tumor (i.e., to address the large and growing number of patients who receive a diagnosis of Tumor of Unknown Origin).
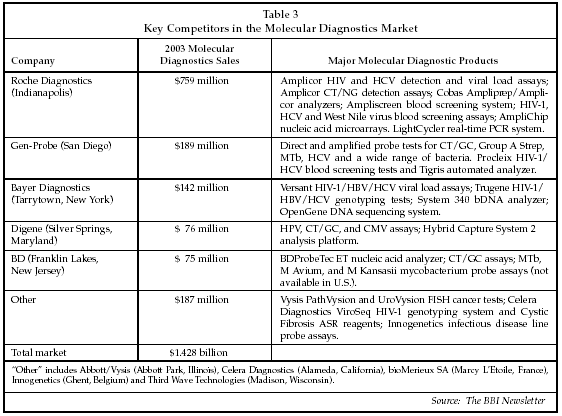
New molecular diagnostic tests for infectious disease also were described at the San Diego Conference. The infectious disease segment continues to represent the majority of the global market and is the primary focus of the major suppliers of molecular diagnostic products, which are listed along with their 2003 global sales and key products in the molecular diagnostics market in Table 3. While some of the market leaders offer products for genetic testing or pharmacogenetics, their revenues in segments other than infectious disease are not significant at present. Some of the suppliers in the "Other" category, such as the Vysis (Downers Grove, Illinois) unit of Abbott Diagnostics (Abbott Park, Illinois), Third Wave Technologies (Madison, Wisconsin) and Celera Diagnostics (Alameda, California), are focusing on the emerging market segments, however. In the infectious disease segment, new tests for viral load and viral genotyping are attracting strong interest, and comprise some of the most rapidly growing subsegments of the market.
A new technology for HIV and HCV viral load testing was described by Mark Holodniy, MD, director of the AIDS Research Center and HIV Clinical Program at Stanford Medical School (Palo Alto, California). The method is targeted at testing of patients in less-developed regions, such as Africa. An absorbent filter, called the Sample Tanker, is a key component of the technology, and allows a 1 mL plasma sample to be preserved during transmit via regular mail from remote regions to a central testing facility in the U.S. or Europe. Holodniy has demonstrated that viral load measurements performed on such samples exhibit quite low variance compared to frozen specimens, and has also been able to automate the sample preparation process once the dried sample is received. For drug resistance assessment, a Virtual Phenotype test method, the VircoTYPE from Virco Diagnostics (Baltimore), can be used which costs about 50% less than conventional phenotyping tests, or about $400 to $600 per test.
Another test discussed by Holodniy that is proving useful in tracking patient response to therapy is a proteomic-mapping assay developed by Zyomyx (Hayward, California). The Zyomyx assay has been shown to track changes in chemokine expression in response to HIV therapy, and the changes in expression are also correlated with patient response. Holodniy said there is a need for new completely automated viral load and genotyping assays for AIDS patient management, as well as less-expensive tests for use in the Third World.
Needs also exist for portable, integrated test systems that can be used not only for infectious disease testing but also in biodefense applications. Companies addressing the development of portable, point-of-care nucleic acid analyzers include Gen-Probe (San Diego), which recently announced a collaboration and licensing agreement with Qualigen (Carlsbad, California) to develop a POC nucleic acid analyzer based on Qualigen's FastPack immunoassay technology.
